RadAR: the new software developed by the Hospital of Prato, IFAC-CNR and the University of Florence
The ongoing 4th Industrial Revolution is also impacting the healthcare, especially in fields such as the bioinformatics, the Internet of Things (IoT), the Artificial Intelligence (AI), the robotics, and etc.
The quantitative analysis of biomedical images, referred to as radiomics, represents an example of the application of AI and IoT tools in the context of precision oncology to provide valuable information on the features or mutations of tumors, and on how to personalize their treatments.
Thanks to RadAR (Radiomics Analysis with R), the software developed by the Researchers and medical physicists from USL Toscana Centro, the Institute of Applied Physics “Nello Carrara” and the University of Florence, it is possible to perform the analysis of radiomic data in a complete and detailed way.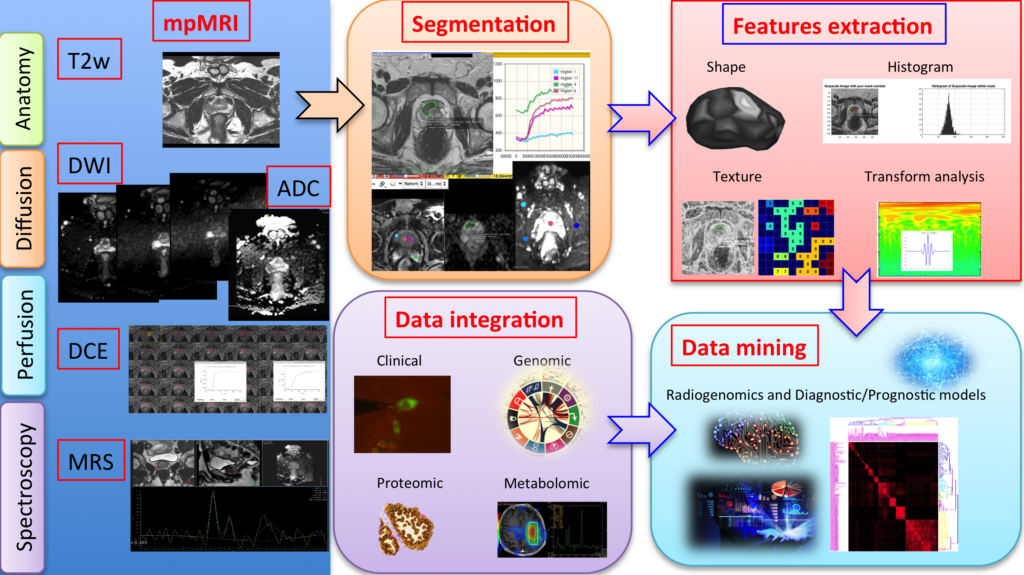
Figure 1. Workflow of Radiomics
Radiological techniques, such as Computed Tomography (CT), Magnetic Resonance Imaging (MRI), and Positron Emission Tomography (PET), are able to generate images of very high quality that in recent years have contributed to improve the diagnosis and monitoring of pathological conditions, especially in the field of oncology. Recently, the results of these exams have started to be analyzed not only as images for visual inspection (qualitatively or semi-qualitatively), but also as a quantitative database.
This approach is very promising, because it allows detecting information that may otherwise be missed by human operators. Radiomics is in fact a new discipline that deals with the extraction of quantitative features from radiological images, which are then processed through analytical and statistical methods. This approach allows for diagnoses that are more accurate, and can also provide valuable information on the prognosis of specific tumors, such as predicting the response to a certain treatment or highlighting the presence of specific genetic and epigenetic alterations.
To demonstrate the reliability of the RadAR software, the researchers used Computed Tomography data from over 850 oncological patients, obtained by open access databases. The new software was open source distributed via the GitHub platform, and it is available for free to all researchers interested in this new discipline. The international journal Cancer Research has published the results of this study, and is available here.
“When we started working on this data, we quickly realized that there was no software dedicated to their analysis and, above all, that there was no consensus on how to perform these analyzes – explains Dr. Matteo Benelli, Head of the Bioinformatics Unit in Prato and coordinator of the study – These techniques generate a large amount of data and pose questions similar to those that we encounter every day in the sector of the bioinformatics and genomics of tumors. Thanks to a great interdisciplinary work, we have developed RadAR, and made it available to the whole scientific community. We therefore hope – continues Matteo Benelli – that our software may in the future facilitate complex analyzes such as radiomics, and contribute to the spread of this new discipline, which may make an important impact on improving the quality of care for cancer patients.”
Researchers Andrea Barucci and Nicola Zoppetti from IFAC-CNR contributed to this work, by evaluating the radiomic features, i.e. the mathematical functions designed to extract quantitative information from radiological images, and then by validating the entire RadAR software package. “Based on a series of clinical images that are available as open source, after the identification of an area corresponding to a certain lesion, we have evaluated its features, such as the distribution of intensity tones, the texture, the shape, and other mathematical functions like the fractal dimension” – They explain – “We have thus created and optimized a pipeline for the extraction of these features, capable of interfacing with the server where clinical images are stored, and pre-processing them if necessary to make them compatible with the standards of the calculation software. In addition to radiological images, the analytical pipeline also requires the so-called ‘segmented images’, i.e. maps where the areas corresponding to the pathological lesion under examination are segmented by the medical doctor. Having these two types of images as input, the new procedure is able to calculate the features of interest and organize the results in a structured way that can be imported into the RadAR software for subsequent statistical analysis.” “The number of features normally extracted from a radiological image is very large” – continues Andrea Barucci – “from a few hundred to many thousands, with parameters that must be set, adapted and tested on the basis of the type of image and of the pathology / tissue under examination. All this requires a high degree of automation of the process, and at the same time a significant extent of control over the whole procedure, in order to avoid introducing errors that may affect the final statistics.”
The study was made possible thanks to financial support to the Bioinformatics Unit of the Prato Hospital by CR Firenze Foundation, and thanks to the collaboration of Sandro Pitigliani Foundation, involved in raising funds to support research carried out in the Oncology Facility in Prato, directed by Dr. Angelo Di Leo, and part of the Department of Oncology department directed by Dr. Luisa Fioretto.
Source: FONDAZIONE CR FIRENZE and CNR


